Kaiser-Guttman Criterion
KGC.RdThis function implements the Kaiser-Guttman criterion (Guttman, 1954; Kaiser, 1960) for determining the number of factors to retain in factor analysis. It is based on the eigenvalues of the correlation matrix of the responses. According to the criterion, factors are retained if their corresponding eigenvalues are greater than 1.
KGC(
response,
fa = "pc",
nfact = 1,
cor.type = "pearson",
use = "pairwise.complete.obs",
vis = TRUE,
plot = TRUE
)Arguments
- response
A required
N×Imatrix or data.frame consisting of the responses ofNindividuals toIitems.- fa
A string that determines the method used to obtain eigenvalues. If 'pc', it represents Principal Component Analysis (PCA); if 'fa', it represents Principal Axis Factoring (a widely used Factor Analysis method; @seealso
factor.analysis; Auerswald & Moshagen, 2019). (Default = 'pc')- nfact
A numeric value that specifies the number of factors to extract, only effective when
fa = 'fa'. (Default = 1)- cor.type
A character string indicating which correlation coefficient (or covariance) is to be computed. One of
"pearson"(default),"kendall", or"spearman". @seealsocor.- use
an optional character string giving a method for computing covariances in the presence of missing values. This must be one of the strings
"everything","all.obs","complete.obs","na.or.complete", or"pairwise.complete.obs"(default). @seealsocor.- vis
A Boolean variable that will print the factor retention results when set to
TRUE, and will not print when set toFALSE. (default =TRUE)- plot
A Boolean variable that will print the KGC plot when set to
TRUE, and will not print it when set toFALSE. @seealsoplot.KGC. (Default =TRUE)
Value
An object of class KGC is a list containing the following components:
- nfact
The number of factors to be retained.
- eigen.value
A vector containing the empirical eigenvalues
References
Guttman, L. (1954). Some necessary conditions for common-factor analysis. Psychometrika, 19, 149–161. http://dx.doi.org/10.1007/BF02289162.
Kaiser, H. F. (1960). The application of electronic computers to factor analysis. Educational and Psychological Measurement, 20, 141–151. http://dx.doi.org/10.1177/001316446002000116.
Examples
library(EFAfactors)
set.seed(123)
##Take the data.bfi dataset as an example.
data(data.bfi)
response <- as.matrix(data.bfi[, 1:25]) ## loading data
response <- na.omit(response) ## Remove samples with NA/missing values
## Transform the scores of reverse-scored items to normal scoring
response[, c(1, 9, 10, 11, 12, 22, 25)] <- 6 - response[, c(1, 9, 10, 11, 12, 22, 25)] + 1
## Run KGC function with default parameters.
# \donttest{
KGC.obj <- KGC(response)
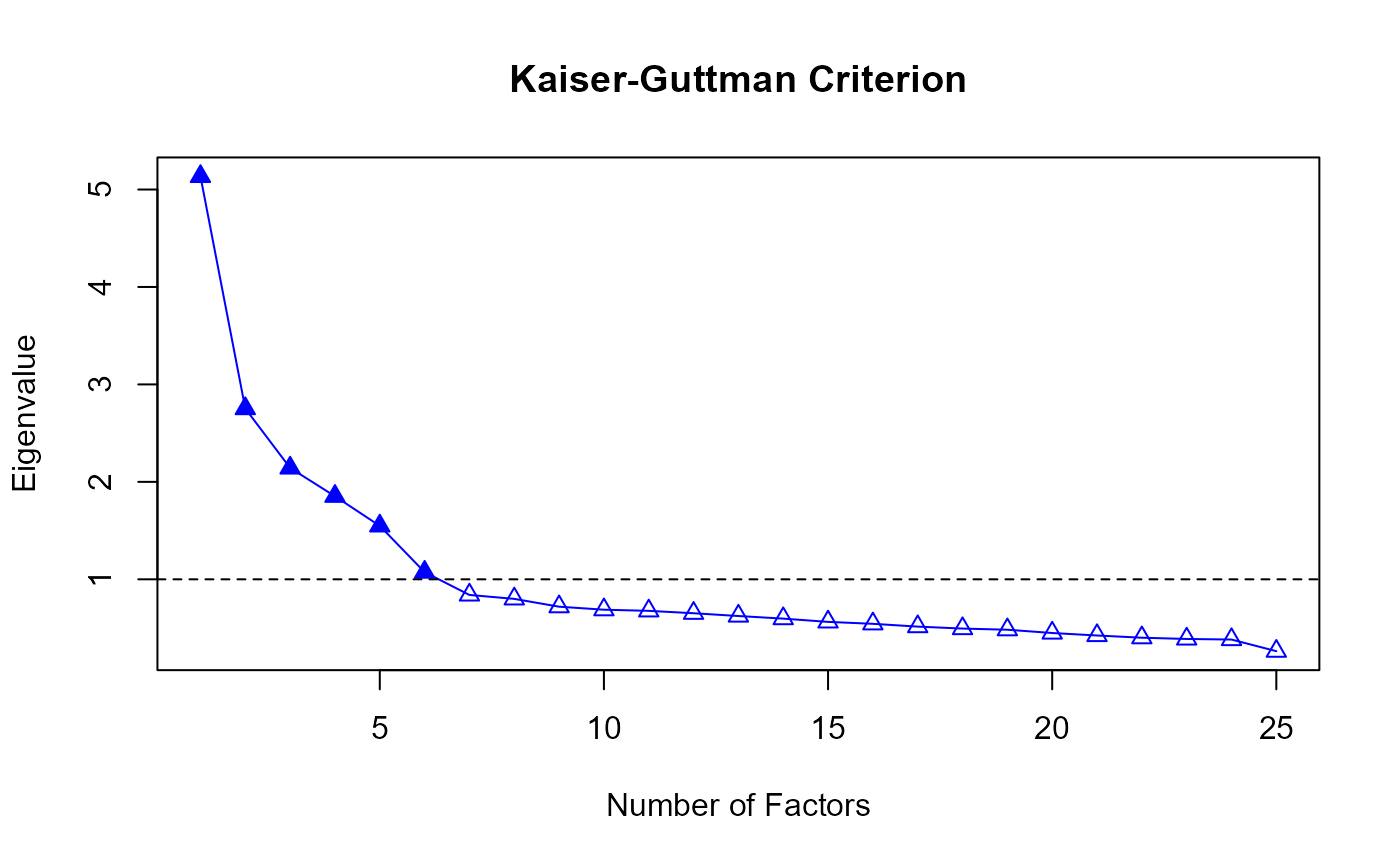 #> The number of factors suggested by KGC is 6 .
print(KGC.obj)
#> The number of factors suggested by KGC is 6 .
plot(KGC.obj)
## Get the eigen.value, eigen.ref and nfact results.
eigen.value <- KGC.obj$eigen.value
nfact <- KGC.obj$nfact
print(eigen.value)
#> [1] 5.1343112 2.7518867 2.1427020 1.8523276 1.5481628 1.0735825 0.8395389
#> [8] 0.7992062 0.7189892 0.6880888 0.6763734 0.6517998 0.6232530 0.5965628
#> [15] 0.5630908 0.5433053 0.5145175 0.4945031 0.4826395 0.4489210 0.4233661
#> [22] 0.4006715 0.3878045 0.3818568 0.2625390
print(nfact)
#> [1] 6
# }
#> The number of factors suggested by KGC is 6 .
print(KGC.obj)
#> The number of factors suggested by KGC is 6 .
plot(KGC.obj)
## Get the eigen.value, eigen.ref and nfact results.
eigen.value <- KGC.obj$eigen.value
nfact <- KGC.obj$nfact
print(eigen.value)
#> [1] 5.1343112 2.7518867 2.1427020 1.8523276 1.5481628 1.0735825 0.8395389
#> [8] 0.7992062 0.7189892 0.6880888 0.6763734 0.6517998 0.6232530 0.5965628
#> [15] 0.5630908 0.5433053 0.5145175 0.4945031 0.4826395 0.4489210 0.4233661
#> [22] 0.4006715 0.3878045 0.3818568 0.2625390
print(nfact)
#> [1] 6
# }