Empirical Kaiser Criterion
EKC.RdThis function will apply the Empirical Kaiser Criterion (Braeken & van Assen, 2017) method to determine the number of factors. The method assumes that the distribution of eigenvalues asymptotically follows a Marcenko-Pastur distribution (Marcenko & Pastur, 1967). It calculates the reference eigenvalues based on this distribution and determines whether to retain a factor by comparing the size of the empirical eigenvalues to the reference eigenvalues.
EKC(
response,
cor.type = "pearson",
use = "pairwise.complete.obs",
vis = TRUE,
plot = TRUE
)Arguments
- response
A required
N×Imatrix or data.frame consisting of the responses ofNindividuals toIitems.- cor.type
A character string indicating which correlation coefficient (or covariance) is to be computed. One of
"pearson"(default),"kendall", or"spearman". @seealsocor.- use
an optional character string giving a method for computing covariances in the presence of missing values. This must be one of the strings
"everything","all.obs","complete.obs","na.or.complete", or"pairwise.complete.obs"(default). @seealsocor.- vis
A Boolean variable that will print the factor retention results when set to
TRUE, and will not print when set toFALSE. (default =TRUE)- plot
A Boolean variable that will print the EKC plot when set to
TRUE, and will not print it when set toFALSE. @seealsoplot.EKC. (Default =TRUE)
Value
An object of class EKC is a list containing the following components:
- nfact
The number of factors to be retained.
- eigen.value
A vector containing the empirical eigenvalues
- eigen.ref
A vector containing the reference eigenvalues
Details
The Empirical Kaiser Criterion (EKC; Auerswald & Moshagen, 2019; Braeken & van Assen, 2017) refines Kaiser-Guttman Criterion by accounting for random sample variations in eigenvalues. At the population level, the EKC is equivalent to the original Kaiser-Guttman Criterion, extracting all factors whose eigenvalues from the correlation matrix are greater than one. However, at the sample level, it adjusts for the distribution of eigenvalues in normally distributed data. Under the null model, the eigenvalue distribution follows the Marčenko-Pastur distribution (Marčenko & Pastur, 1967) asymptotically. The upper bound of this distribution serves as the reference eigenvalue for the first eigenvalue \(\lambda\), so
$$\lambda_{1,ref} = \left( 1 + \sqrt{\frac{I}{N}} \right)^2$$
, which is determined by N individuals and I items. For subsequent eigenvalues, adjustments are made based on the variance explained by previous factors. The j-th reference eigenvalue is:
$$\lambda_{j,ref} = \max \left[ \frac{I - \sum_{i=0}^{j-1} \lambda_i}{I - j + 1} \left( 1 + \sqrt{\frac{I}{N}} \right)^2, 1 \right]$$
The j-th reference eigenvalue is reduced according to the magnitude of earlier eigenvalues since higher previous values mean less unexplained variance remains. As in the original Kaiser-Guttman Criterion, the reference eigenvalue cannot drop below one.
$$F = \sum_{i=1}^{I} I(\lambda_i > \lambda_{i,ref})$$
Here, \( F \) represents the number of factors determined by the EKC, and \(I(\cdot)\) is the indicator function, which equals 1 when the condition is true, and 0 otherwise.
References
Auerswald, M., & Moshagen, M. (2019). How to determine the number of factors to retain in exploratory factor analysis: A comparison of extraction methods under realistic conditions. Psychological methods, 24(4), 468-491. https://doi.org/10.1037/met0000200.
Braeken, J., & van Assen, M. A. L. M. (2017). An empirical Kaiser criterion. Psychological methods, 22(3), 450-466. https://doi.org/10.1037/met0000074.
Marcˇenko, V. A., & Pastur, L. A. (1967). Distribution of eigenvalues for some sets of random matrices. Mathematics of the USSR-Sbornik, 1, 457–483. http://dx.doi.org/10.1070/SM1967v001n04ABEH001994
Examples
library(EFAfactors)
set.seed(123)
##Take the data.bfi dataset as an example.
data(data.bfi)
response <- as.matrix(data.bfi[, 1:25]) ## loading data
response <- na.omit(response) ## Remove samples with NA/missing values
## Transform the scores of reverse-scored items to normal scoring
response[, c(1, 9, 10, 11, 12, 22, 25)] <- 6 - response[, c(1, 9, 10, 11, 12, 22, 25)] + 1
## Run EKC function with default parameters.
# \donttest{
EKC.obj <- EKC(response)
#> The number of factors suggested by EKC is 6 .
 print(EKC.obj)
#> The number of factors suggested by EKC is 6 .
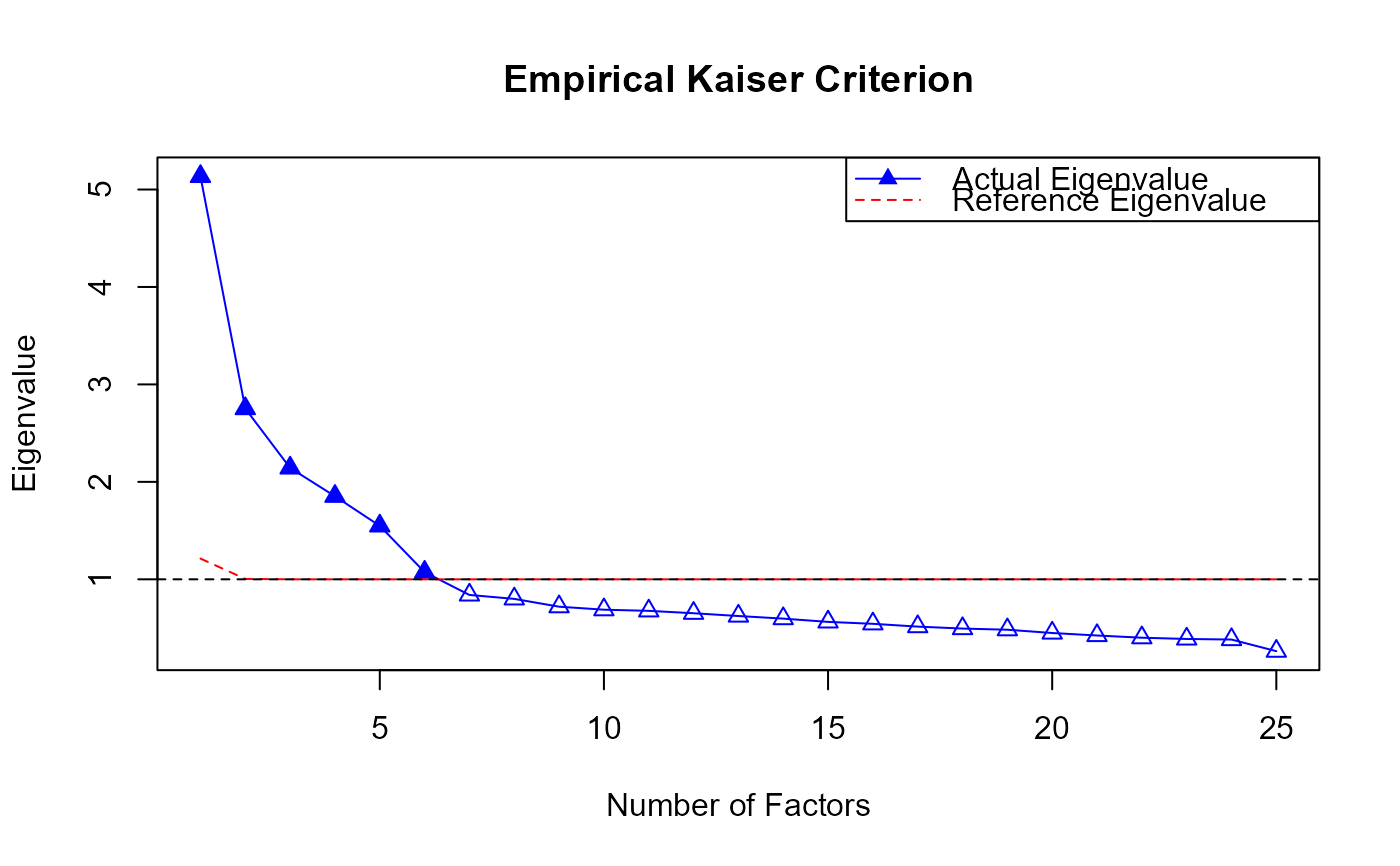
plot(EKC.obj)
## Get the eigen.value, eigen.ref and nfact results.
eigen.value <- EKC.obj$eigen.value
eigen.ref <- EKC.obj$eigen.ref
nfact <- EKC.obj$nfact
print(eigen.value)
#> [1] 5.1343112 2.7518867 2.1427020 1.8523276 1.5481628 1.0735825 0.8395389
#> [8] 0.7992062 0.7189892 0.6880888 0.6763734 0.6517998 0.6232530 0.5965628
#> [15] 0.5630908 0.5433053 0.5145175 0.4945031 0.4826395 0.4489210 0.4233661
#> [22] 0.4006715 0.3878045 0.3818568 0.2625390
print(eigen.ref)
#> [1] 1.212873 1.003940 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000
#> [9] 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000
#> [17] 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000
#> [25] 1.000000
print(nfact)
#> [1] 6
# }
print(EKC.obj)
#> The number of factors suggested by EKC is 6 .
plot(EKC.obj)
## Get the eigen.value, eigen.ref and nfact results.
eigen.value <- EKC.obj$eigen.value
eigen.ref <- EKC.obj$eigen.ref
nfact <- EKC.obj$nfact
print(eigen.value)
#> [1] 5.1343112 2.7518867 2.1427020 1.8523276 1.5481628 1.0735825 0.8395389
#> [8] 0.7992062 0.7189892 0.6880888 0.6763734 0.6517998 0.6232530 0.5965628
#> [15] 0.5630908 0.5433053 0.5145175 0.4945031 0.4826395 0.4489210 0.4233661
#> [22] 0.4006715 0.3878045 0.3818568 0.2625390
print(eigen.ref)
#> [1] 1.212873 1.003940 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000
#> [9] 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000
#> [17] 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000
#> [25] 1.000000
print(nfact)
#> [1] 6
# }